Searching and acquiring IRIS Level 2 data
Contents
1. Searching and acquiring IRIS Level 2 data#
Looking for IRIS Level 2 data is quite simple thanks to the tools available in the IRIS web page. There in the right-hand column, Quick Links, we can find the IRIS Level 2 data search tool. In the IRIS tutorial web page the user can find many other interesting tutorials such as the “Acquiring IRIS data” tutorial or Section 3.2 of A User’s Guide to IRIS Data Retrieval, Reduction & Analysis. We refer the reader to these tutorials as a great way to learn how to look for IRIS Level 2 data using the search tools designed for this goal.
One of the advantages of using the IRIS Level 2 data search tool lies in its ability to make complex search using the size of the field-of-view scanned, exposure time, location, available SJI data in specific channels, and in coordination with other observatories. This powerful tool allows one to recover the events found with these, or other, conditions, in either simpler or complex queries. After the user has filled the search fields with their search conditions and clicked Search button, the observations (or events) found that comply with the desired conditions will be shown in the right-hand panel. Note that the list of events actually satisfying the search conditions may be larger than the list showed in this panel. To avoid that, the user can increase the maximum number of events to be displayed by changing the value in the Limit drop list widget next to Search button.
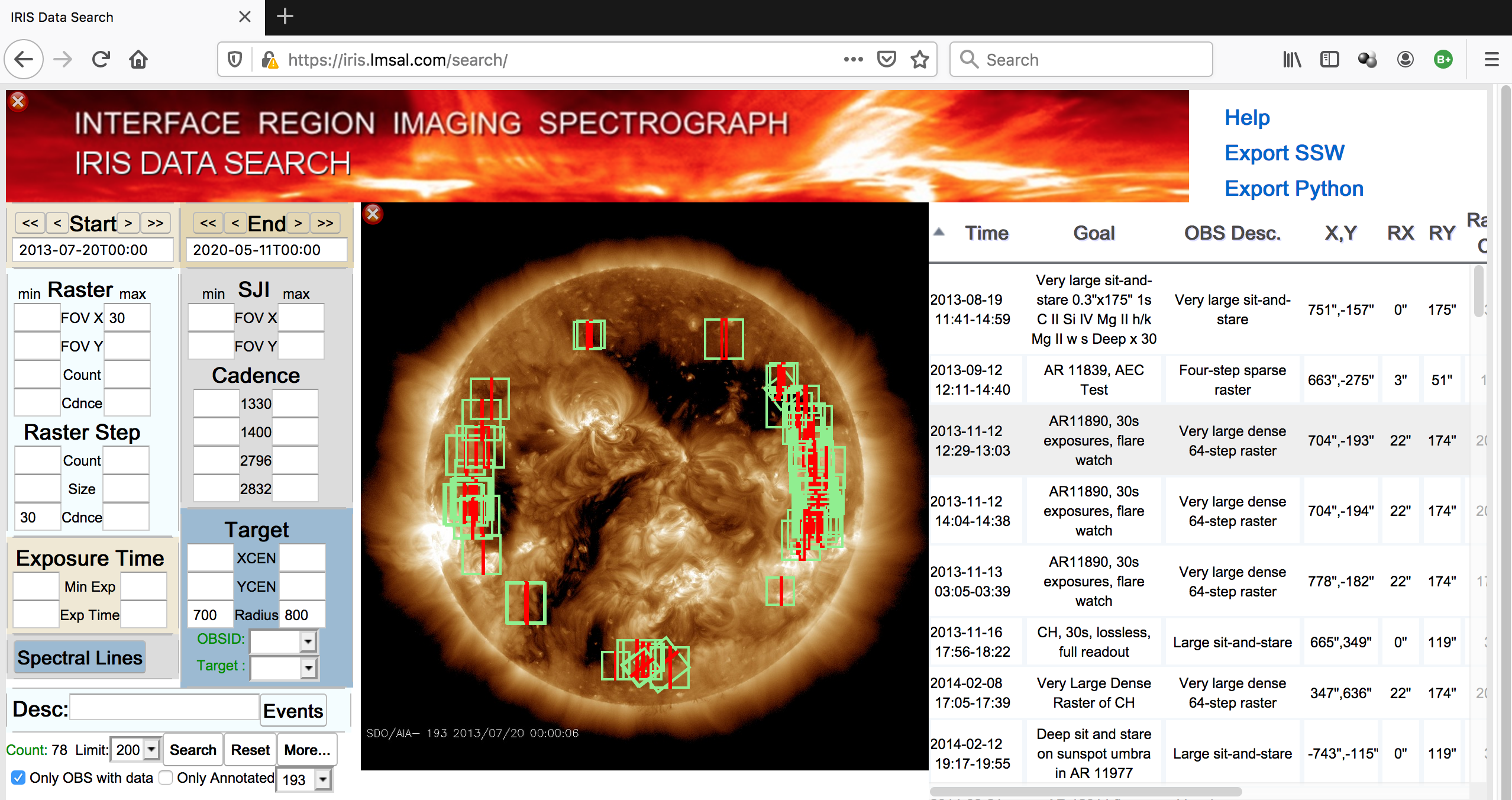
Fig. 1.1 The IRIS Level 2 data search tool with a complex search parameters that found 16 events in the whole IRIS database. Clicking in the Export Python link will open a new tab (or window) with the Python commands needed to download the data.#
Fig. 1.1 shows the IRIS L2 data search tool after a relatively complex search.
In the latest version of the IRIS Level 2 data search tool, there is a link to recover the Heliophysics Coverage Registry (HCR) record(s) that satisfy the search conditions using Python. This link is shown as Export Python. After clicking on that link, a new window or tab with an automatically-generated text will show some commands similar to these ones:
1>>> from iris_lmsalpy import hcr2fits
2>>> query_text = 'https://www.lmsal.com/hek/hcr?cmd=search-events3&outputformat=json&startTime=2016-01-14T00:00&stopTime=2016-01-15T00:00&minnumRasterSteps=320&hasData=true&minxCen=550&limit=200'
3>>> list_urls = hcr2fits.get_fits(query_text)
The last command will download and decompress data from NOAA AR 12480 that we will use in this guide.
The hcr2fits code was created to download the compressed IRIS Level 2 data files found by the IRIS Level 2 data search tool.
The input is query_text (line 3), a string automatically generated by this tool.
Several keywords allow the behavior of this routine to be changed and you can check the function documentation.
Note
hcr2fits assumes that gunzip, tar, and wget are properly installed in the user’s machine.
If that is not the case, you can install them by executing the following commands in your shell session:
conda install tar
conda install wget
The other choice is to install them system wide using your operating system’s package manager or homebrew equivalent.
The total size of the downloaded and decompressed files is \(\approx 1.9 Gbytes\), and they are:
$ ls -lh
... 166M Dec 3 02:15 iris_l2_20160114_230409_3630008076_SJI_1330_t000.fits
... 166M Dec 3 02:15 iris_l2_20160114_230409_3630008076_SJI_1400_t000.fits
... 166M Dec 3 02:15 iris_l2_20160114_230409_3630008076_SJI_2796_t000.fits
... 166M Dec 3 02:15 iris_l2_20160114_230409_3630008076_SJI_2832_t000.fits
... 1.3G Jul 31 2017 iris_l2_20160114_230409_3630008076_raster_t000_r00000.fits
After copying and pasting the previous Python commands into your Python session, the code will do the following:
Generate a list of links to the compressed IRIS Level 2 data located on the IRIS data server
Request permission to download the found data by using
wgetDownload the compressed data if the permission is given by the IRIS data server
After all the compressed data are downloaded, decompress the downloaded data by using
tarandgunzip.
Using the get_fits command, all these step are done automatically, if the connection to the IRIS data server is working properly and the space in the user’s machine is large enough to write and decompress the data.
Note that the number of the total found events is generally much smaller than the number of files to be written in the destination folder.
To guarantee better understanding and performance of the downloading process, the code accepts several keywords that allow the user to:
output_dir: set the destination folder where the data will be written and decompressed. Default isNone, that means, the data will be downloaded to the current working directory, i.e.,./.decompress: set/unset the automatic decompression of the downloaded data. Default isTrue, i.e. automatic decompression is doneraster_only: download only the raster files, i.e.,iris_l2_*raster*fitsfiles. Default isFalsesji_only: download only the SJI files, i.e.,iris_l2_*SJI*fitsfiles, Default isFalsetext_files: generate two shell script files (iris_wget_query.shandiris_decompress_query.sh) to manually download the data. Ifdecompress = True, it creates the fileiris_decompress_query.sh, which allows the user to decompress the data manually in the system command line as a script. The user can comment out some download/decompress tasks in these files, for example if the list of found files is too long, by preceding the commands in the text files with a`#. Default value isFalse.
Warning
The following examples are intended to show the capability of hcr2fits to download and decompress a large set of data.
Therefore, they require a stable and fast connection to the internet with enough space in the system to store the data.
It is not necessary to run these commands for the normal continuity of this tutorial.
These commands are here presented only for educational purposes.
We may be interested in to look at a large set of IRIS Level 2 data. This is the case of the search shown in Fig. 1.1. In these cases, it may be useful to generate shell scripts files that contain the commands needed to download and decompress the data at the user’s discretion:
# It will create two shell scripts to manually download and decompress the SJI files.
>>> query_text = "https://www.lmsal.com/hek/hcr?cmd=search-events3&outputformat=json&startTime=2013-07-20T00:00&stopTime=2020-04-04T00:00&minhpcRadius=300&maxhpcRadius=400&minexpMin=5&maxrasterFOVX=16&maxnumRasterSteps=5&maxcadMeanPlannedSJI2796=20&hasData=true&limit=200"
>>> list_urls = hcr2fits.get_fits(query_text, sji_only = True, text_files = True)
>>> print('{} SJI files have been found'.format(len(list_urls)))
Requesting the query...
Creating iris_wget_query.sh
Creating iris_decompress_query.sh
39 SJI files have been found
If the data need a large space to be stored in a location different than the current working directory (default behavior), we can set the output with the keyword output_dir:
# It will download both the raster and the SJI files into the directory
# /scratch/asainz/IRIS_data/, but it will not decompress them.
>>> query_text = 'https://www.lmsal.com/hek/hcr?cmd=search-events3&outputformat=json&startTime=2013-07-20T00:00&stopTime=2020-04-04T00:00&minhpcRadius=300&maxhpcRadius=400&minexpMin=5&maxrasterFOVX=16&maxnumRasterSteps=5&maxcadMeanPlannedSJI2796=20&hasData=true&limit=200'
>>> list_urls = hcr2fits.get_fits(query_text, output_dir = '/scratch/asainz/IRIS_data/', decompress = False)
Requesting the query...
Downloading the file http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2019/05/08/20190508_162936_3803109418/iris_l2_20190508_162936_3803109418_raster.tar.gz into /scratch/asainz/IRIS_data/ (#1 of 53) ...
URL transformed to HTTPS due to an HSTS policy
--2020-04-08 22:04:10-- https://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2019/05/08/20190508_162936_3803109418/iris_l2_20190508_162936_3803109418_raster.tar.gz
Resolving www.lmsal.com (www.lmsal.com)... 166.21.250.149
Connecting to www.lmsal.com (www.lmsal.com)|166.21.250.149|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 860673031 (821M) [application/x-gzip]
Saving to: '/scratch/asainz/IRIS_data/iris_l2_20190508_162936_3803109418_raster.tar.gz'
iris_l2_20190508_16 1%[> ] 15.37M 866KB/s eta 16m 1s
...
many other files are downloaded and decompressed
Both the code and the shell scripts generated by the former show progress status information during their execution.
1.1. Saving your search (and other variables)#
The output of hcr2fits is a list containing the URLs to the found compressed IRIS Level 2 data files (list_urls in the examples):
>>> for i, url in enumerate(list_urls):
print(i, url)
0 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2019/08/09/20190809_100336_3602088422/iris_l2_20190809_100336_3602088422_SJI_2796_t000.fits.gz
1 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2019/08/09/20190809_100336_3602088422/iris_l2_20190809_100336_3602088422_SJI_1400_t000.fits.gz
2 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2019/08/09/20190809_100336_3602088422/iris_l2_20190809_100336_3602088422_SJI_2832_t000.fits.gz
3 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2016/07/04/20160704_100551_3680258322/iris_l2_20160704_100551_3680258322_SJI_1330_t000.fits.gz
4 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2016/07/04/20160704_100551_3680258322/iris_l2_20160704_100551_3680258322_SJI_2796_t000.fits.gz
5 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2016/07/04/20160704_100551_3680258322/iris_l2_20160704_100551_3680258322_SJI_2832_t000.fits.gz
6 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2019/05/08/20190508_162936_3803109418/iris_l2_20190508_162936_3803109418_SJI_2796_t000.fits.gz
7 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2019/05/08/20190508_162936_3803109418/iris_l2_20190508_162936_3803109418_SJI_1400_t000.fits.gz
8 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2018/10/09/20181009_154453_3620028413/iris_l2_20181009_154453_3620028413_SJI_2796_t000.fits.gz
9 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2018/10/09/20181009_154453_3620028413/iris_l2_20181009_154453_3620028413_SJI_1400_t000.fits.gz
10 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2018/10/09/20181009_154453_3620028413/iris_l2_20181009_154453_3620028413_SJI_2832_t000.fits.gz
11 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2019/05/11/20190511_162936_3803109418/iris_l2_20190511_162936_3803109418_SJI_2796_t000.fits.gz
12 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2019/05/11/20190511_162936_3803109418/iris_l2_20190511_162936_3803109418_SJI_1400_t000.fits.gz
13 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2016/07/02/20160702_092921_3680258322/iris_l2_20160702_092921_3680258322_SJI_1330_t000.fits.gz
14 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2016/07/02/20160702_092921_3680258322/iris_l2_20160702_092921_3680258322_SJI_2796_t000.fits.gz
15 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2016/07/02/20160702_092921_3680258322/iris_l2_20160702_092921_3680258322_SJI_2832_t000.fits.gz
16 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2016/05/15/20160515_072919_3640008423/iris_l2_20160515_072919_3640008423_SJI_2796_t000.fits.gz
17 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2016/05/15/20160515_072919_3640008423/iris_l2_20160515_072919_3640008423_SJI_1400_t000.fits.gz
18 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2016/05/15/20160515_072919_3640008423/iris_l2_20160515_072919_3640008423_SJI_2832_t000.fits.gz
19 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2013/11/07/20131107_071938_3860259419/iris_l2_20131107_071938_3860259419_SJI_2796_t000.fits.gz
20 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2013/11/07/20131107_071938_3860259419/iris_l2_20131107_071938_3860259419_SJI_1400_t000.fits.gz
21 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2014/10/05/20141005_114150_3860359362/iris_l2_20141005_114150_3860359362_SJI_1330_t000.fits.gz
22 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2014/10/05/20141005_114150_3860359362/iris_l2_20141005_114150_3860359362_SJI_2796_t000.fits.gz
23 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2014/09/24/20140924_112436_3860359362/iris_l2_20140924_112436_3860359362_SJI_1330_t000.fits.gz
24 http://www.lmsal.com/solarsoft/irisa/data/level2_compressed/2014/09/24/20140924_112436_3860359362/iris_l2_20140924_112436_3860359362_SJI_2796_t000.fits.gz
25 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/11/19/20141119_190039_3800259365/iris_l2_20141119_190039_3800259365_SJI_1330_t000.fits.gz
26 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/11/19/20141119_190039_3800259365/iris_l2_20141119_190039_3800259365_SJI_2796_t000.fits.gz
27 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/11/19/20141119_203839_3800259365/iris_l2_20141119_203839_3800259365_SJI_1330_t000.fits.gz
28 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/11/19/20141119_203839_3800259365/iris_l2_20141119_203839_3800259365_SJI_2796_t000.fits.gz
29 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/09/27/20140927_112936_3860359362/iris_l2_20140927_112936_3860359362_SJI_1330_t000.fits.gz
30 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/09/27/20140927_112936_3860359362/iris_l2_20140927_112936_3860359362_SJI_2796_t000.fits.gz
31 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/09/26/20140926_110936_3860359362/iris_l2_20140926_110936_3860359362_SJI_1330_t000.fits.gz
32 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/09/26/20140926_110936_3860359362/iris_l2_20140926_110936_3860359362_SJI_2796_t000.fits.gz
33 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/05/24/20140524_142104_3820109371/iris_l2_20140524_142104_3820109371_SJI_1330_t000.fits.gz
34 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/05/24/20140524_142104_3820109371/iris_l2_20140524_142104_3820109371_SJI_2796_t000.fits.gz
35 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/05/28/20140528_161436_3820109371/iris_l2_20140528_161436_3820109371_SJI_1330_t000.fits.gz
36 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2014/05/28/20140528_161436_3820109371/iris_l2_20140528_161436_3820109371_SJI_2796_t000.fits.gz
37 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2013/11/07/20131107_094438_3860259419/iris_l2_20131107_094438_3860259419_SJI_2796_t000.fits.gz
38 http://www.lmsal.com/solarsoft/irisa/data_lmsal/level2_compressed/2013/11/07/20131107_094438_3860259419/iris_l2_20131107_094438_3860259419_SJI_1400_t000.fits.gz
We can keep a record of our search by saving this information with saveall.
You can save almost any kind of object (variable) with saveall as far as the object does not have a method (function) associated.
That means, you can save variables such as arrays, list, dictionaries and others, or more complex objects with this kind of attributes.
It is not possible to save either methods (functions) or objects with associated methods with saveall.
This function is a wrapper code developed by A. Sainz Dalda based on the Pickle and joblib libraries.
It follows a similar command structure that save in IDL, i.e., saveall.save(filename, var1, var2, ..., varN).
An important constraint of saveall is that the save method has to be a 1-line command.
Usually that is not a problem if the list of variables is not too long.
# Let's save this list. We use saveall function to this aim
>>> from iris_lmsalpy import saveall as sv
>>> file_info_mysearch = 'myIRISsearch_ring_700_800.jbl.gz'
>>> sv.save(file_info_mysearch, list_urls, query_text)
Saving...
list_urls
query_text
... in myIRISsearch_ring_700_800.jbl.gz
The code shows the variable(s) saved in the destination file. If the showed variables does not match with the variables asked to be saved that means something went wrong.
If save detects that the file used to store the objects already exists, then the code will ask you to continue with the writing process:
>>> sv.save(file_info_mysearch, list_urls, query_text)
# Answer 'n'
File myIRISsearch_ring_700_800.jbl.gz exists. Do you want to overwrite it? Y/[n] n
Nothing has been done.
To avoid this safety question you can set the keyword force as True:
>>> sv.save(file_info_mysearch, list_urls, query_text, force = True)
Saving...
list_urls
query_text
... in myIRISsearch_ring_700_800.jbl.gz
It is very easy to recover the saved objects, list_urls and query_text in this example, from the file that stores them:
>>> file_info_mysearch = 'myIRISsearch_ring_700_800.jbl.gz'
>>> aux = sv.load(file_info_mysearch)
Loading joblib file... myIRISsearch_ring_700_800.jbl.gz
Suggested commands:
list_urls = aux['list_urls']
query_text = aux['query_text']
del aux
The variable types are:
list_urls : <class 'list'>
query_text : <class 'str'>
saveall.load method (or sv.load in the example) returns a dictionary object containing the variables saved in the file as the keys of the returned dictionary.
As you can see, a list of suggested commands are prompted by this method.
We suggest to copy and paste these commands to restore the variables saved with their original names into the current namespace, i.e., the current Python session if saveall.load is executed in the command line, or the global or local namespace if it is executed inside of a Python code.
>>> list_urls = aux['list_urls']
>>> query_text = aux['query_text']
>>> del aux
>>> type(list_urls)
list
>>> type(query_text)
str
